A Genomic Atlas
Building a Genomic Atlas for Pol III
Currently I aim to understand the genetic landscape for RNA Polymerase III. My research project aims to build a multi-tissue and cancer atlas of Pol III, uncovering context-specific transcriptional expansion and identifying genes implicated in disease progression. In order to achieve this, I have applied different statistical methods to define significance on genes based on their chromatin accessibility profiles.
Maybe you might have not heard too much about Pol III, but the truth is, that this is an amazing enzyme which is totally understudied (Lots of attention of Pol II). Let’s share some fun facts about Pol III:
- Pol III is in charge of transcribing a plethora of small non coding RNAs, one of them being tRNAs.
- Pol III is the largest RNA polymerase protein complex and is comprised of multiple subunits that are either Pol III-specific or otherwise shared with the other polymerases.
- One of these subunits is POLR3, which has two paralogous subunits: POLR3G which is highly expressed during early development and subsequently attenuated during differentiation and POLR3GL which is expressed at later stages of development and required for long-term survival in vivo.
- In a cancer context POLR3G expression reemerges. Therefore it is linked with poor outcomes in patients.
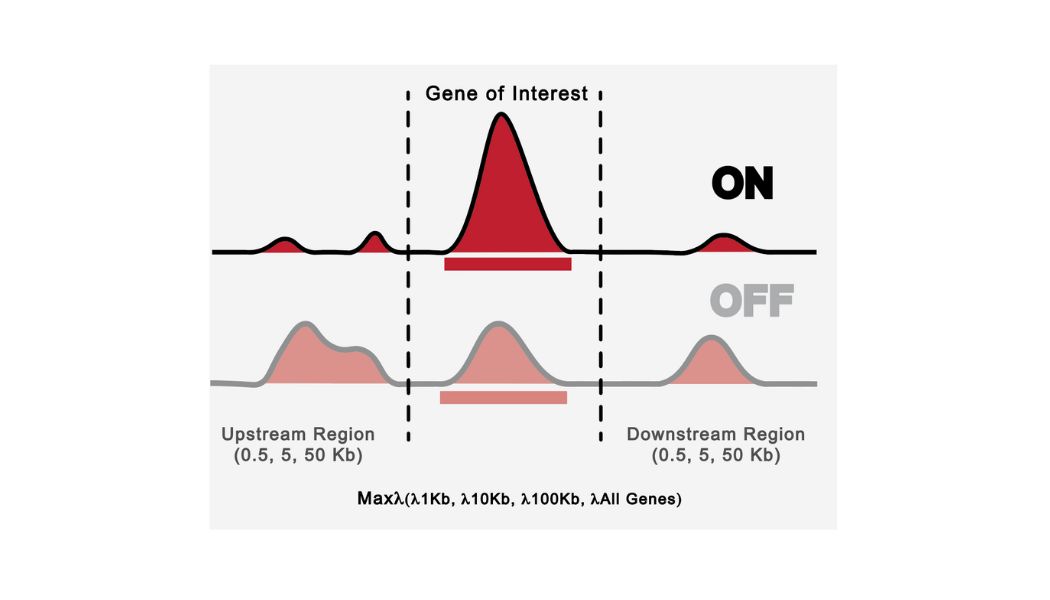
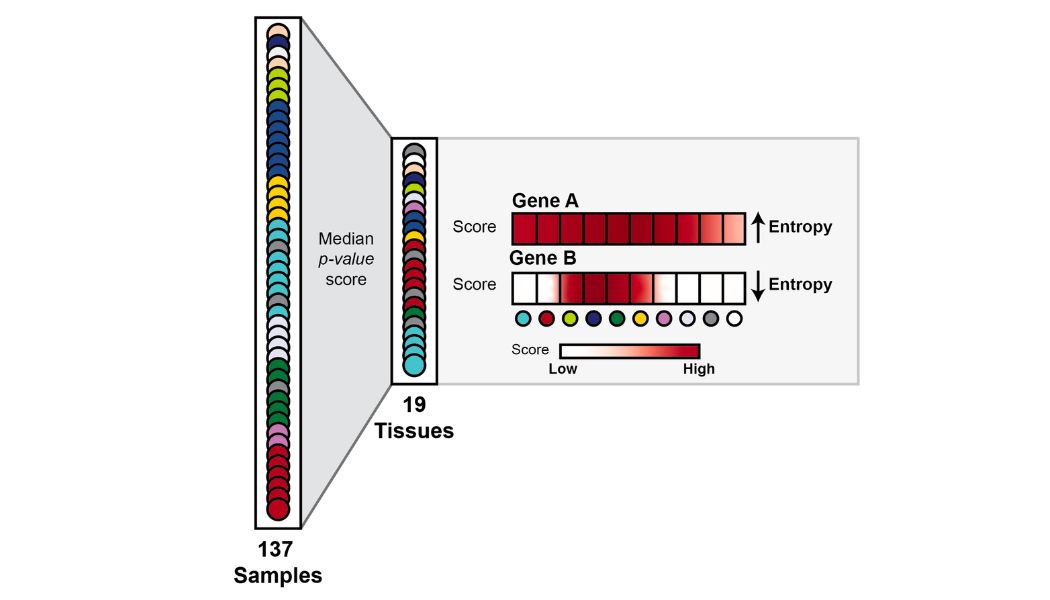
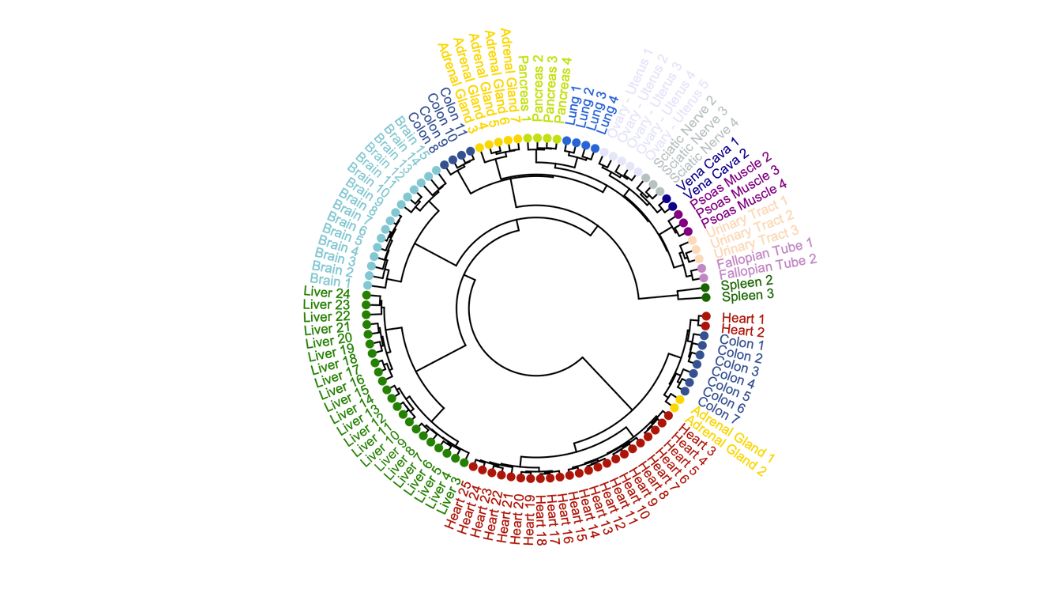
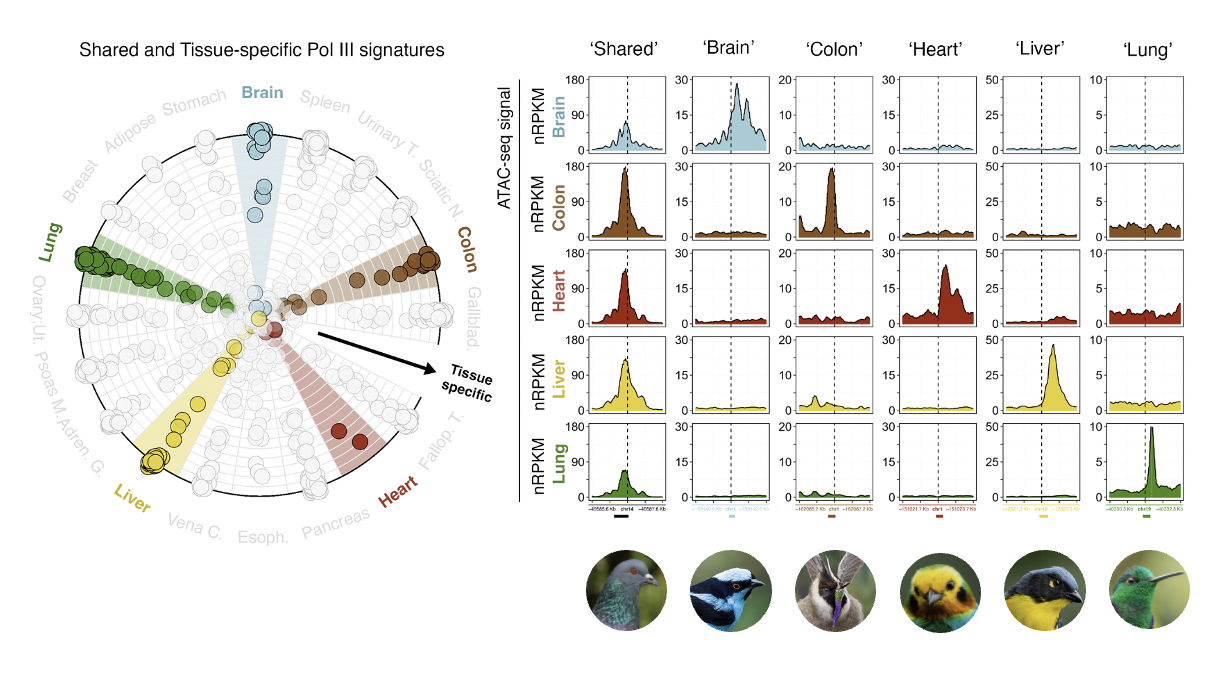
I have developed an interesting method to determining how likely a gene is to be accessible at the chromatin level by analyzing several ATAC-Seq files. I have been able to define different groups of Pol III genes and explored different epigenetic features for these groups.
Some key findings about this project:
- Pol III chromatin accessibility profiles are highly tissue/cancer specific , with this in mind, I have been able to define a list of highly uniform and less uniform (specific) genes across tissues
- In the cancer context, I have been able to observe an expansion of the Pol III transcriptome when compared to Tissues.
- There are a large set of epigenetic modifications across different groups of genes that help to explain their tissue/cancer diversification.
Throughout this project I have been able to integrate different types of genomic data (Chip-Seq, DamID-Seq, ATAC-Seq, RNA-Seq, WGBS, etc). Moreover I am trying to build an R Package for feature dominance visualization motivated by the challenges faced trying to optimally represent data across multiple variables effectively. I am working on this manuscript! Wait for it!